myDGR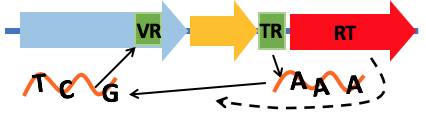
Click here to Check out Demonstration examples
Input
- Input sequences (in FASTA format) Users can upload a sequence file or paste the sequence(s) in the submission page. All sequences must be in the FASTA format. To ensure that every sequence has a unique ID, we will check and assign an ID number to each sequence. Special characters (including white space, dot and comma) will be removed from the sequences. However, if a sequence contains illegal letters (e.g., numbers), the job will be terminated.
- Input GFF file (optional) As an option, users can also provide gene information in GFF (General Feature Format) format. Otherwise, myDGR will call FragGeneScan to predict protein coding genes. In the GFF file, the information of contig/genome ID, gene location and gene translation direction must be provided.
- Metagenome option: If the metagenome box is checked, myDGR will apply an extra step to group DGRs it finds according to the RT gene similarity, as a metagenome may contain similar DGR systems. Also myDGR will skip the step of finding remote target genes if metagenome box is checked. Check out the result of myDGR results for a toy metagenome
Output
- Visual summary shows all the important features a DGR system has, including the reverse transcriptase (RT, shown as red arrow below), putative target gene (blue), putative accessory gene (orange), the
TR and VR pair (shown as green boxes), and the hairpin structure (shown as a red hairpin in the figure below)
- MyDGR provides iteractive exploration (implemented using SVG) of the DGR systems. Users can move mouse over to different components of the DGR systems to see the details and domain composition of the domains. Interactive exploration works on most web browsers including Chrome, Firefox and Safari. However, if it doesn't work somehow for your favoriate brower -- please consider trying a different one if you want to use this functionality. [see an example of visual summary and domain visualization]
- The hallmark feature of the DGR system is the TR repeat and VR repeat, which are similar, except the A postions in the TR repeat. An alignment of the TR-VR repeats
can show where the substitutions happen in the pair, as shown below

- In the cases where there are multiple target genes subject to diversification by a DGR system, you can click on the MSA in the summary page to see the multiple alignment of the TR and VR segments.

[try it] - Accessory genes Accessory genes are often found in the immediate neighborhood of the core DGR components (i.e., the RT, TR and the nearby VR-containing target gene). MyDGR extracts adjacent two genes in both directions of identified RT gene, and compare their protein sequences against the HMMs of domains built from previously identified accessory genes using hmmscan search. The protein predicted to contain one of these domains is reported as the putative accessory gene. However, accessory genes are poorly conserved, and thus some of them might be missed by the similarity searches. If that happens, the visual summary of the DGR systems produced by myDGR will provide a convenient way for users to manually annotate potential accessory genes; and users can download the protein sequences of the genes close to RT gene (by following the genes next to RT link).
- Target proteins and their domain composition
The VR-encoding regions in target proteins can be clustered into five main groups (CLec1, CLec2, Clec3, Ig1 and Ig2; see paper). We assign the VR regions predicted by myDGR into one of these classes using hmmscan. We further search for putative Pfam domains using hmmscan for predicted target proteins.
The figure belows shows the domain composition predicted for a target gene, with domains shown as boxes, the VR-encoding domain (CLec1 in this case) highlighted in purple, and other domains shown in either green (if hmmscan E-value < 0.001) or gray (otherwise).
In particular, the VR-encoding region (337-381) in the target protein is indicated by a yellow bar below the protein.The figures below show the weblogo of the five main classes of the VR-encoding regions (the multiple alignments of the domains were downloaded from here).- CLec1
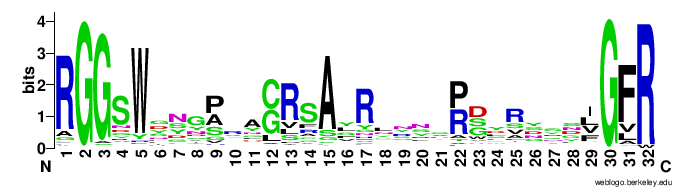
- CLec2
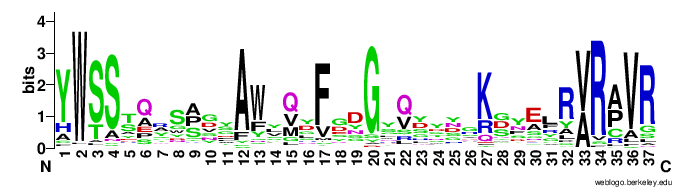
- CLec3
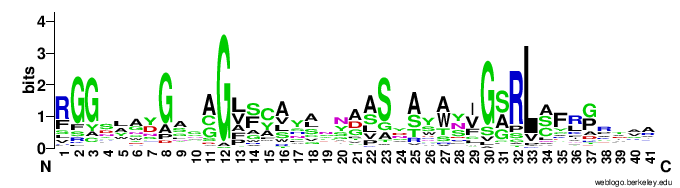
- Ig1
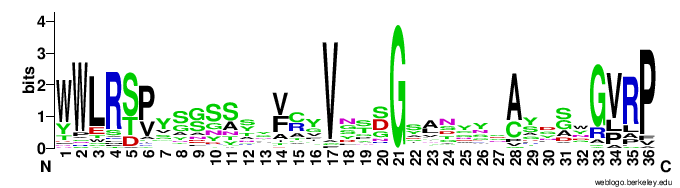
- Ig2

- CLec1
- Text files for download: gff file with all the annotated features (RT, TR/VR, hairpin, target gene), targe gene, reverse transcriptase, and so on.
- If the input is a metagenome (and the user selected the "Metagenome" option") myDGR provides an additional option of grouping its predicted DGR systems according to the RT similarity.
Check out the result of myDGR results for a toy metagenome. In this case, two clusters of DGR systems were detected--the first one contains one instance, and the second cluster contains two instances.
- Check out Demonstration examples
- For more information about Reverse Transcriptases associated with DGR systems, checkout myRT
Please contact us if you have questions: Contact us